About the LBMP Genome Database
About the LBMP Genome Database
Material selected for generation of reference genome includes plants of SP70-1143 sugarcane genotype grown in soil. Young leaves were collected and the genomic DNA was purified from isolated nuclei of -2 leaves.
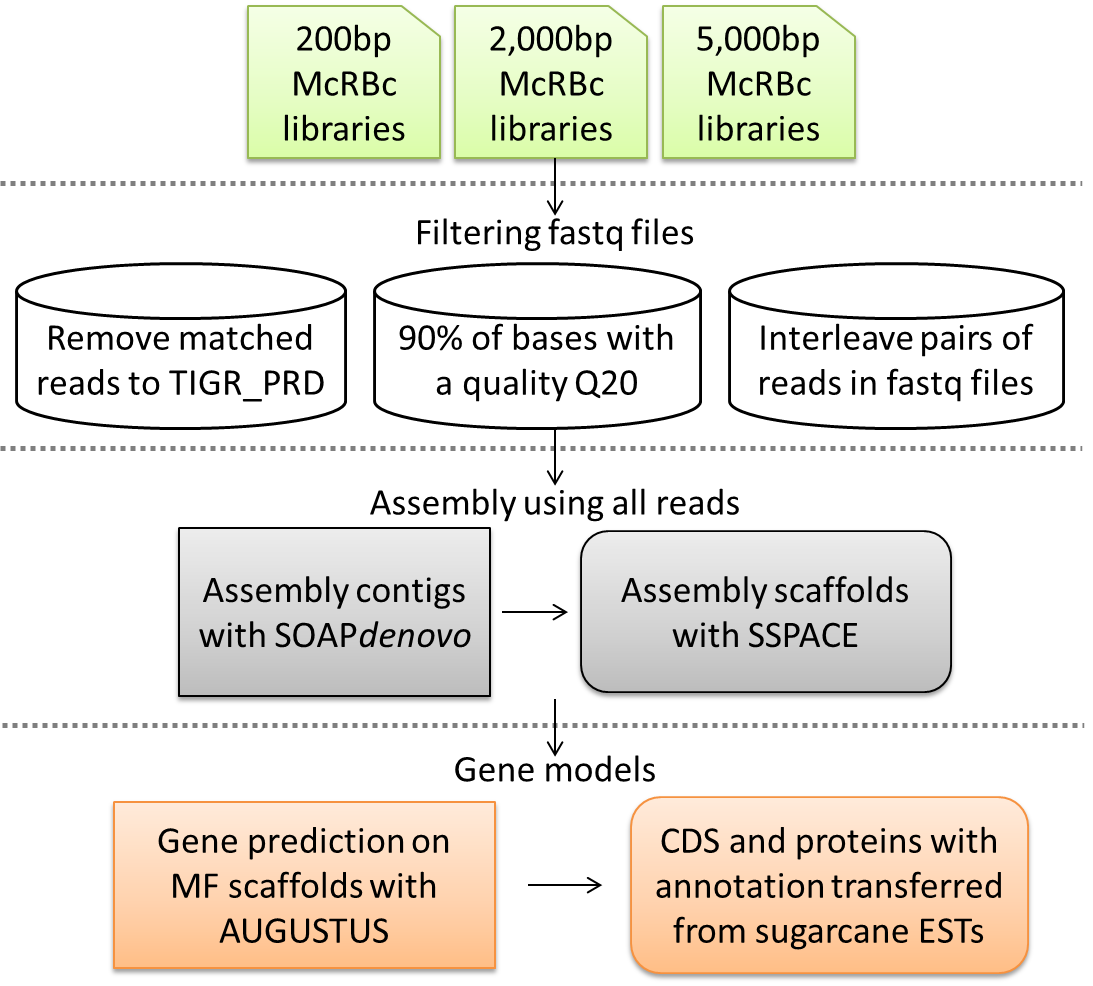
The MF (Methyl filtered) technology was applied with Illumina short reads sequencing to generate a set of sequences from the hypomethylated portion of sugarcane genome. The MF libraries containing inserts of 200bp size were constructed and sequenced at Cold Spring Harbor Laboratory on Illumina GAII machine using the paired-end 50 cycle protocol. Two mate-pair libraries were constructed with selection of genomic DNA sheared at 2,000 and 5,000 bp, followed by McrBC treatment. The libraries containing inserts of 2 and 5 kb sizes were sequenced at Fasteris Life Sciences SA (Plan-les-Ouates, Switzerland) on HiSeq2000 machine using the paired-end 100 cycle protocol. Construction and sequencing of MF libraries resulted in 286,452,478 reads obtained. The raw fastq files have been submitted to the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession no. SRP023506.
The quality-filtered reads from 3 McrBC digested libraries were quality filtered (90% of bases having a base quality greater or equal than 20) and after that, cleaned for repetitive sequences by sequence match to TIGR_Gramineae_Repeats.v3.3_0_0. The remaining reads were shuffled on each paired library and first assembled into contigs with SOAPdenovo using a range of kmer sizes (25, 31, 37, 41, 43, 49). The best kmer size (k=49) was chose based on size include N, size without N, mean size and number of contigs over 500 bp. The pre-assembled contigs were scaffolded by using SSPACE_basic v.2.0. Only scaffolds above 200bp were considered. The sequences generated were identified by the ID number and their length correlated to it (e.g. scaffold1475|size6679).
The sugarcane CDS and proteins from SOAPdenovo+SSPACE scaffolds were predicted using AUGUSTUS. The existing gene models of the closest organism was used (i.e. Rhizopus oryzae) to predict genes on sugarcane scaffolds. The annotation of sugarcane CDS and proteins were transferred from the EST annotation using best hit between ESTs aligned with predicted CDS and protein sequences from sugarcane scaffolds. The predicted sugarcane CDS and proteins with annotation kept the scaffold identification by the ID number and the length correlated to the scaffold (e.g. scaffold1475|size6679).
LBMP Genome database workflow